On April 15, Professor GAO Shaorong and Professor ZHANG Yong from the School of Life Science and Technology published, in collaboration with students in their research team, a paper entitled dynamic nucleosome organization after fertilization reveals regulatory factors for mouse zygotic genome activation online in Cell Research. From the perspective of nucleosome position on chromatin, this research reveals the dynamic process and regulation mechanism of chromatin reorganization of parents within 12 hours after fertilization, emphasizes the significance of histone-based chromatin state regulation for normal development, and unveils the regulation secret at the beginning of life.

The paper published in Cell Research
For eukaryotes like mammals, DNA, as genetic material, needs to be wound around histones to form the basic structural unit of chromatin - nucleosomes. Nucleosomes, also the primary structure of chromatin, are the basis for the subsequent agglutination of chromatin to form a more complex spatial structure, and affect the transcriptional activity of the genome. The recording and embodiment of genetic information by chromosomes in cells is like a knot in ancient civilization, and DNA molecules are like a long rope, which is the material basis of information recording. Nucleosome is the most basic knot on the rope. It is the basic structure for the recording and transmission of genetic information in cells. It has different distribution patterns in different types of cells. In the process of spermatogenesis, the histone in the nuclear corpuscle is replaced by the unique protamine of sperm, so that the chromatin can be compressed together and present a highly agglutinated state. In the initial fertilization process of life, this highly compressed chromatin will disintegrate, protamine will be replaced by histone, and the nucleosome structure in the general state will be re-formed, so as to complete the remodeling of chromatin and create the foundation for zygotic genome activation (ZGA). However, for a long time, it has been unclear how these changes come about and develop at the genomic level and what factors regulate them.
What we did was figure out how to untie the tangled rope in the gamete, tie a new knot again, and get ready for the transmission of genetic information, said GAO Shaorong. However, this process changes very fast and needs to be refined by hour. It is also necessary to distinguish the genomes of parents from different sources. More importantly, it is necessary to solve the miniaturization of detection technology.
The research team has developed a new technology (ULI-MNase-seq, patented) to detect the arrangement of genomic nucleosomes on low starting amount (even single cell), and developed a process of analysis NEPTUNE specifically for nucleosome data. According to GAO, they separated the male and female pronucleus manually at 9 consecutive time points from 0.5 to 12 hours after fertilization through micromanipulation, and detected the nucleosome arrangement of these pronucleus and gametophyte cells, thus completing the fine description of the dynamic remodeling of nuclear topics 12 hours before fertilization for the first time. It was found out that the parent genome can complete the replacement of protamine to histone about an hour after fertilization, and the establishment of nucleosome deleted region (NDR) related to transcriptional activity in male prokaryotic was also faster.
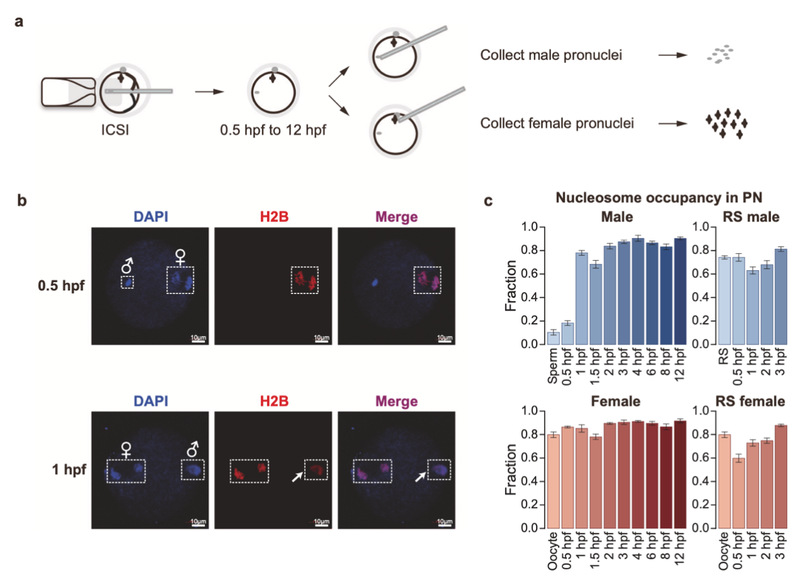
Figure 1. Nucleosome arrangement patterns during fertilization
The research team explored the factors affecting the early nucleosome arrangement. It was found that the content of GC in DNA sequence (the ratio of guanine to cytosine) has a significant effect on the occupancy of nucleosome, and the acetylation modification on histone may regulate the formation of nucleosome NDR. In addition, the research team discovered that some transcription factors would gradually replace nucleosomes during fertilization, bind to specific sequence regions of the genome, and may be involved in the formation of nucleosome NDR and the transcriptional activation of specific genes in a wider promoter region. The researchers revealed two transcription factors Mlx and Rfx1 respectively, and found that Mlx could regulate the occurrence of ZGA at zygote stage, while Rfx1 could significantly affect ZGA at 2-cell stage.
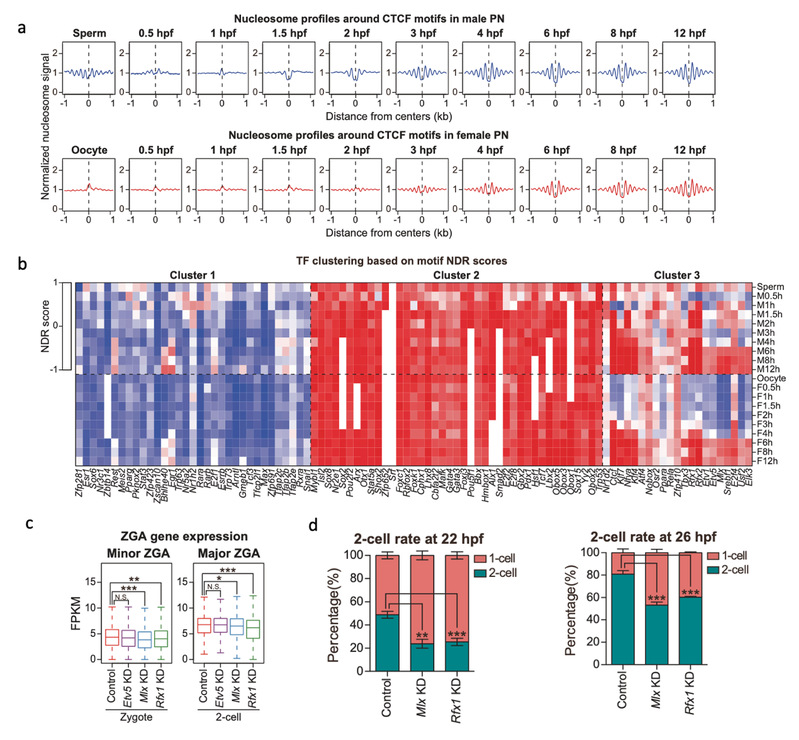
Figure 2. Transcription factors affecting nucleosome arrangement and ZGA
Our research has drawn a high-resolution map of nucleosome arrangement during male and female prokaryotic fertilization in mice for the first time, revealed the establishment pattern of nucleosome arrangement, and predicted new regulatory factors. The nucleosome body weight arrangement process in fertilization is an important link in the exploration of the regulation mystery in the initial stage of life, which is not only very important for developmental biology, but also very important for assisted reproduction and human reproductive health. GAO Shaorong said that this research provides strong support for people to understand the transmission of epigenetic modifications between generations and the regulation of gene expression during embryonic development.
WANG Chenfei, CHEN Chuan, LIU Xiaoyu and LI Zhen of Tongji University are the co-first authors of the paper, and GAO Shaorong, ZHANG Yong and GAO Yawei are the corresponding authors of the paper. The research was supported by projects funded by the Ministry of Science and Technology and the Shanghai Science and Technology Commission.